Generating synthetic electronic health record (EHR) data using denoising diffusion probabilistic models
Our recent work on 'Synthesizing Mixed-type Electronic Health Records using Diffusion Models' is now available on arXiv! 📢
Note: This article is best viewed in ‘light mode’!
Challenge
The latest advances in machine learning and artificial intelligence (AI/ML) has opened up new possibilities to improve healthcare outcomes by harnessing the large volumes of data contained in electronic health records (EHRs). However, due to the sensitive nature of the information contained in EHRs, strict data sharing requirements and the need to preserve patient privacy has hindered the development and deployment of AI/ML models across healthcare settings.
Solution
Generating synthetic patient data, instead, offers a promising solution to mitigate these risks, although creating realistic data is often very tricky and still an open challenge. In our recent work, led by Dr. Taha Ceritli, we have explored how a new state-of-the-art type of generative model, known as Denoising Diffusion Probabilistic Models (DDPMs), could generate more realistic synthetic EHR data beyond existing techniqus, such as well-known Generative Adversarial Networks (GANs).
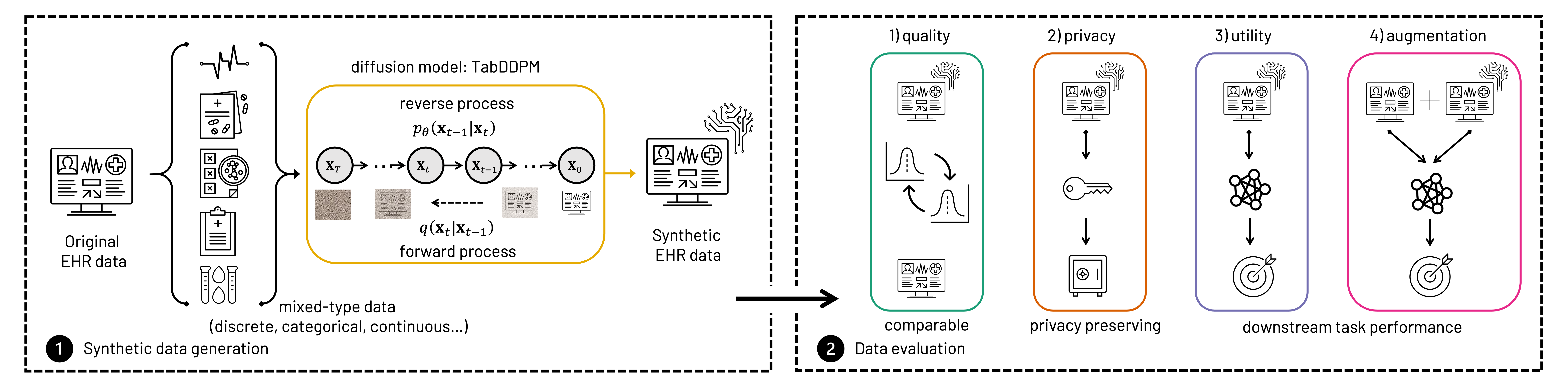
An overview of the TabDDPM pipeline: step (1) demonstrates the synthetic data generation process, followed by step (2) where the synthetic data is evaluated based on quality, privacy, utility, and augmentation performance.
Denoising Diffusion Probabilistic Models (DDPMs)
Diffusion models are inspired by non-equilibrium thermodynamics, and they learn to generate synthetic data through denoising. Learning by denoising consists of two processes, forwards and reverse, each of which is a Markov Chain:
Forward process
In the forward process, random noise is added to the orginal data in a series of time steps \((t_1, t_2, \ldots, t_n)\), following \(q(\mathbf{x}_t|\mathbf{x}_{t-1})\), where \(q(\cdot)\) is a Markovian process that produces the corrupted samples at each \(t\). Samples at each time step are drawn from a Gaussian distribution, where the mean of the distribution is conditioned on the sample at the previous time step, and the variance of the distribution follows a fixed schedule. At the end of the forward process, the samples become pure noise.
Reverse process
During the reverse process, we try to remove the added noise at every time step, beginning with the pure noise distribution (the last step of the forward process). During the reverse process we can only approximate \(q(\mathbf{x}_{t−1}|\mathbf{x}_t)\), which we do using the conditional probabilities \(p_{\theta}(\mathbf{x}_{t−1}|\mathbf{x}_t)\), as estimated from our neural network. We then try to denoise each sample backwards from \((t_n, t_{n-1}, \ldots, t_1)\) to the original data.
Evaluation
Our diffusion model, known as TabDDPM due to its flexibility incorporating tabular- and mixed-type data, was compared on four evaluation criteria:
- Data quality: can similar and comparable data be generated?
- Data privacy: Can the the privacy of original patients be preserved?
- Data utility: Can synthesized data be useful for downstream predictive models?
- Data augmentation: Can combining original and synthetic data strengthen the capacity of downstream predictive models?
We found that TabDDPM outperformed many of the state-of-the-art methods on benchmark datasets, except for privacy, highlighting the difficult balance that needs to be maintained when synthesizing EHR. Our the abilty to generate more realistic clinical data comes with the cost of too closly mimicing the unique and confidential characteristics of the original patients.
We’re not there yet, but we hope that our promising results motivates future work exploring how diffusion models could generate realistic healthcare data, without compromising on the privacy considerations of patients.
Feedback
This project is still evolving and we welcome any feedback. If you have comments or suggestions, feel free to reach out to myself or Dr. Taha Ceritli. Congratulations to all co-authors, Taha Ceritli, Ghadeer Ghosheh, Vinod Kumar Chauhan, Tingting Zhu and David Clifton.
Reference
If you use our work, please consider citing:
@article{ceritli2023synthesizing,
title = {Synthesizing Mixed-type Electronic Health Records using Diffusion Models},
author = {Ceritli, Taha and Ghosheh, Ghadeer O. and Chauhan, Vinod Kumar and Zhu, Tingting and Creagh, Andrew P. and Clifton, David A.},
doi = {https://doi.org/10.48550/arxiv.2302.14679},
url = {https://arxiv.org/abs/2302.14679},
publisher = {arXiv},
year = {2023},
}